new, course, bioinformatics
Diving into Data: How We Used COVID-19 to Teach Bioinformatics
Pavlin Poličar
Aug 10, 2024
Throughout the years that our lab has been working with data, teaching machine learning courses, and running data mining workshops, it has become evident that the best way to learn about any particular topic is to get your hands dirty and just dive into the data. This is why we developed Orange in the first place; to provide an easy-to-use interface that makes exploring data a piece of cake.
Several years ago, we decided to apply this same line of thinking to teaching our university students about molecular biology and bioinformatics. The COVID-19 pandemic was well underway and affecting nearly everyone on the planet. It was certainly affecting all our master’s students at the University of Ljubljana in Slovenia. We asked ourselves: “Could we somehow use this virus to teach our students about molecular biology and bioinformatics? Surely, our students would be motivated to learn about the virus causing the ongoing pandemic!”
And so, we went to the drawing board and completely redesigned our Introduction to Bioinformatics course, taught to Master’s level Computer Science students at our faculty. The result was a completely new set of lab work assignments centered around the exploration and understanding of the SARS-CoV-2 virus. Students would attend lectures, where they would learn about the theory of molecular biology and bioinformatics algorithms, and then solidify this knowledge during their take-home lab work. Students would not only implement the algorithms they learned about during lectures, but also apply them on a real-world problem. In our case, this real-world problem was the SARS-CoV-2 virus. The assignments would guide students through the same steps that scientists took to understand the virus in high-profile publications. They would learn the fundamental elements of bioinformatic analysis hands-on, covering ORF identification, protein translation, sequence alignment, functional annotation, phylogenetic inference, identifying mutations, recombination analysis, and gene-expression data analysis. Our tentative course schedule is illustrated in the figure below.
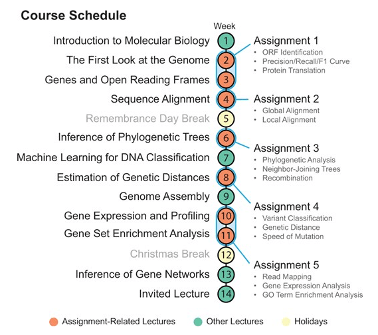
We have been running this course for several years now, doing our best to improve the assignments every year. Each year, we ask students for feedback, and, each year, the feedback has been overwhelmingly positive. This is perhaps surprising since the days of the strict COVID lockdowns are far behind us, but it seems that the virus still manages to capture the attention of our students.
Given this success, we decided to share our course with the broader academic community. Earlier this summer, Pavlin Poličar traveled to Montreal and presented this course at the International Conference on Intelligent Systems for Molecular Biology (ISMB). You can read the full paper titled “Teaching bioinformatics through the analysis of SARS-CoV-2: project-based training for computer science students” in Bioinformatics and access the assignments on GitHub. We also provide instructor notes to guide educators through the logistics of setting up the course.


Courses like Introduction to Bioinformatics serve as wonderful testing grounds for our various ideas about how to effectively teach complex topics like machine learning and bioinformatics. When successful, these ideas sometimes make their way into Orange. Developing visual programming tools for complex topics such as these democratizes these tools, making them available to the more general audience, with perhaps limited or no experience in programming and software development.
